5 U.S. and Global Temperatures
We start our Python and visualization journey by
- Focusing on various Climate Indicators from EPA, primarily from the U.S.
- Using data from EPA
- Relying on the insights generated from these graphs.
We chose to first focus on temperature as the ten most recent years are the warmest years on record.
We can see the headlines in the newspapers and in the media on how intense heat is over the past few years. We just need to step outside in the Summer to feel it! It’s hot :( 🌡️☠️☠️☠️
Heat can exacerbate drought, and hot, dry conditions can in turn create wildfire conditions.
We are using data from EPA to reproduce some of their graphs in their climate indicators.
We do so for two reasons
- As an introductory chapter, we want to make sure that the data is easy to get and analyze and the code is as simple as it can be
- We can compare our graphs with the graphs from EPA to make sure that we are able to reproduce them accurately. We can correct our code in case we find that we are not able to match the graphs from the EPA website
- By reproducing the same graphs as EPA climate indicators, we can learn the same insights and refer to the EPA website without extensively discussing them here.
Remember, our work flow is as follows
Let us start with the concepts!
We need to measure temperature. That is easy!
As we learn from our science class, temperature is measured using Fahrenheit (°F) in the U.S. In this scale, the freezing point of water is 32°F, and the boiling point is 212°F.
In many countries across the world that use metric system, it is measured in Celsius (°C). The scale sets the freezing point of water at 0°C and the boiling point at 100°C under standard atmospheric pressure (212°F).
We use both of them in the book as we visualize both temperatures in the U.S. and across the globe. Also, some of the research papers where we source data present the data in Celsius only.
We need
- A credible source of data
- A way to get data using Python. We don’t want to manually enter all this data :)
As you may have noticed, there are thousands of data sources about climate. There are strong opinions on climate change and different people may interpret the same data differently. The underlying data may not be the same or may have been constructed differently. How do we know which ones to trust and which ones to not trust?
There are many excellent sources of data. In this introductory chapter, we make use of the exact same data that EPA uses for their climate indicators. For each graph on EPA climate indicators website https://www.epa.gov/climate-indicators/climate-change-indicators-united-states-fifth-edition and https://www.epa.gov/climate-indicators/view-indicators
5.1 Historical Temperatures in the U.S. from EPA
Let us start with the EPA data and graphs as they are easier to access and interpret. We can compare our graphs with EPA graphs to make sure that we are on the right path. We can also rely on the EPA’s explanations to understand the findings from these graphs.
EPA has a wealth of climate data at its Climate Indicators website
Information the temperature indicator is available at EPA Temperature Indicator
The page has
- A visualization showing “Temperatures in the Contiguous 48 States, 1901–2023”
- Key Points
- Background
- About the Indicator
- About the Data
- Technical Documentation
Let us start with 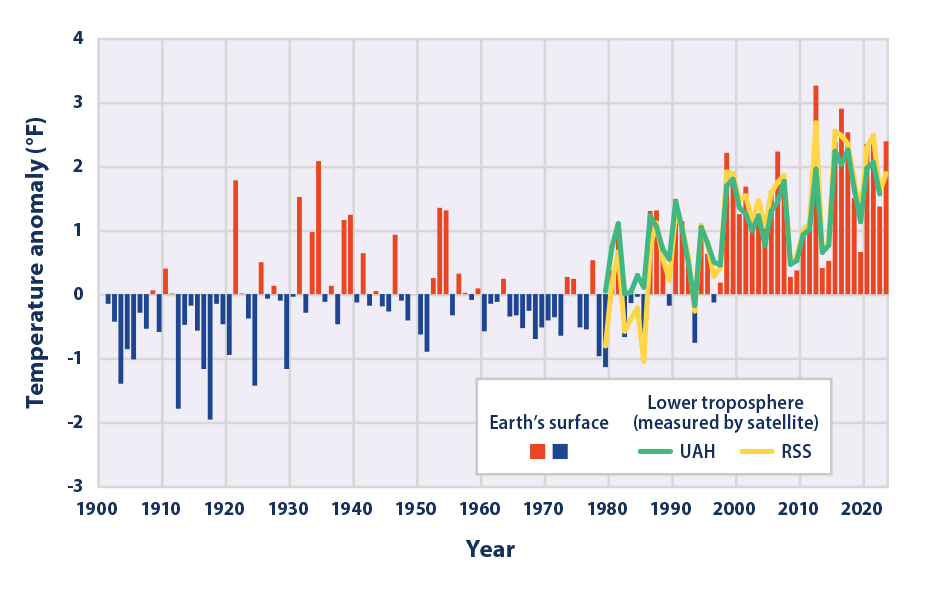
This graph looks interesting! It shows that there recent decades have been very hot as compared to the last hundred or so years.
How do we create this graph? Where we do get the data? How do we arrange the data? How do we create this graph and interpret it?
Before, creating our own visualization, we would encourage the readers to read the entire page as it explains the data, why this indicator is important and the underlying technical documentation. We will refer to some of these later when we try to understand what the graphs are telling us.
We almost always start with importing the necessary libraries. We would extensively use pandas (see our chapter on tools and installing pandas )
By importing pandas with the import statement, we have the all the tools and code in the pandas library available to us. We don’t need to reinvent the wheel and write everything from scratch.
For example, it allows us to read the data from an Excel file or a website. There are many more powerful tools available in pandas such as various data structures and functions that we will use later to clean, transform, analyze, and visualize.
by the way, we gave pd as a shortcut to the pandas library. Instead of using the full name, we can just use pd from now.
Where is the data? How do we get it?
 ,
,
If we notice the visualization, we notice at the left bottom there is a small icon with grids. If we hover the cursor over the icon, we can see the link to a CSV data file. The link is https://www.epa.gov/system/files/other-files/2024-06/temperature_fig-1.csv
Now we have the link, we see that it is a CSV file that can be opened in a text editor or spreadsheet software like Microsoft Excel or Google Sheets.
A CSV (Comma-Separated Values) file is a plain text file that stores tabular data (data organized in rows and columns) in a simple and lightweight format. Each line in a CSV file represents a row, and the values within each row are separated by commas. CSV files are commonly used for data storage and exchange between different software applications.
The data available on the EPA website shows annual anomalies, or differences, compared with the average temperature from 1901 to 2000.
It is always a good idea to open the file and check it as not all CSV files are in the same format, or they may have different headers or footer text.
Let us directly read the data using pandas and print to screen.
import pandas as pd
url = "https://www.epa.gov/system/files/other-files/2022-07/temperature_fig-1.csv"
df = pd.read_csv(url, encoding="ISO-8859-1")
df| Figure 1. Temperatures in the Contiguous 48 States, 1901-2021 | Unnamed: 1 | Unnamed: 2 | Unnamed: 3 | |
|---|---|---|---|---|
| 0 | Source: EPA's Climate Change Indicators in the... | NaN | NaN | NaN |
| 1 | Data source: NOAA, 2022 | NaN | NaN | NaN |
| 2 | Web update: July 2022 | NaN | NaN | NaN |
| 3 | Units: °F | NaN | NaN | NaN |
| 4 | NaN | NaN | NaN | NaN |
| ... | ... | ... | ... | ... |
| 122 | 2017 | 2.53 | 2.219906977 | 2.365113953 |
| 123 | 2018 | 1.5 | 1.610906977 | 1.803213953 |
| 124 | 2019 | 0.66 | 1.094906977 | 1.197363953 |
| 125 | 2020 | 2.35 | 1.937906977 | 2.297463953 |
| 126 | 2021 | 2.49 | 2.024906977 | 2.496663953 |
127 rows × 4 columns
What is the code doing?
- The first imports the pandas library and gives it the alias pd that we can refer to later
- The second line of code assigns a URL to the variable url. This URL points to a CSV file that contains temperature data. We don’t need to do it. We could have directly used the URL
- the third line of code attempts to read this CSV file into a data frame
Note that we are getting the csv. Notice that it’s a link and not downloaded. Since we are going to be using many files throughout this chapter, it’s easier to use the link.
But it is a good idea to assign to a variable and use the variable name later. In case we need to change the URL link, we just need to change this one line.
The second line of code uses the pd.read_csv() function from the Pandas library to read the CSV file specified by the url variable.
Why did we need “encoding=”ISO-8859-1”?
Let us try without it and see what happens.
url = "https://www.epa.gov/system/files/other-files/2022-07/temperature_fig-1.csv"
df = pd.read_csv(url) We will immediately see that we get an error.
UnicodeDecodeError Traceback (most recent call last) 2 url = “https://www.epa.gov/system/files/other-files/2022-07/temperature_fig-1.csv” #<1> —-> 3 df = pd.read_csv(url) #<2> 4
UnicodeDecodeError: ‘utf-8’ codec can’t decode byte 0xb0 in position 228: invalid start byte
The encoding=“ISO-8859-1” parameter is used to specify the character encoding of the CSV file. This is necessary because CSV files can be encoded in different ways, and specifying the correct encoding ensures that the data is read correctly.
The result of reading the CSV file is stored in the variable df. df is a DataFrame object in pandas.
pandas data frame: Data Frame is a two-dimensional tabular data structure provided by the Pandas library.
- Finally, the code outputs the contents of the df DataFrame by simply calling df. This will display the DataFrame in the output pane of the IDE.
To summarize, this code reads temperature data from a CSV file located at the specified URL, stores it in a DataFrame, and then displays the DataFrame in the output pane.
Then, we are making a new DataFrame, df, and reading the dataset. We are using the encoding function because without it (you can try and see) we are getting an error. But why does it look like that?
We notice a few things about the printed data frame
- All the variables named: Figure 1. “Temperatures in the Contiguous 48 States, 1901-2021”, “Unnamed: 1”, “Unnamed: 2”, “Unnamed: 3”.
- There are NaN values for some of the rows.
NaN stands for “Not a Number” and is a special value in pandas that represents missing or undefined data. NaN values can occur in a pandas DataFrame when there is no available value for a particular cell or when the data is not present or cannot be represented as a number.
It depends on the context whether we want to drop the values, set them to zero or treat them differently. A missing value is not always the same as a zero value and can give wrong insights if we treat them the same.
Let us fix these issues!
The first 6 rows seems to include some details in text format about the source of the data are not necessary, so let’s get rid of them.
We have made one change from the previous code. We added “skiprows = 6” in the line where we read the CSV file from the URL.
What does it do?
This is a function call to the read_csv function provided by pandas. It takes several arguments:
url: This is the URL or file path of the CSV file you want to read from the EPA website
skiprows=6: This argument tells pandas to skip the first 6 rows of the CSV file. We are skipping the header information or other irrelevant rows at the beginning.
encoding=“ISO-8859-1”: This argument specifies the character encoding of the CSV file. In this case, it is set to “ISO-8859-1”, which is a common encoding for text files.
Let us see the output
| Year | Earth's surface | Lower troposphere (measured by satellite) (UAH) | Lower troposphere (measured by satellite) (RSS) | |
|---|---|---|---|---|
| 0 | 1901 | -0.15 | NaN | NaN |
| 1 | 1902 | -0.43 | NaN | NaN |
| 2 | 1903 | -1.40 | NaN | NaN |
| 3 | 1904 | -0.86 | NaN | NaN |
| 4 | 1905 | -1.02 | NaN | NaN |
| ... | ... | ... | ... | ... |
| 116 | 2017 | 2.53 | 2.219907 | 2.365114 |
| 117 | 2018 | 1.50 | 1.610907 | 1.803214 |
| 118 | 2019 | 0.66 | 1.094907 | 1.197364 |
| 119 | 2020 | 2.35 | 1.937907 | 2.297464 |
| 120 | 2021 | 2.49 | 2.024907 | 2.496664 |
121 rows × 4 columns
Now we can see the variables or columns are
- ‘Year’
- ‘Earth’s surface’
- ‘Lower troposphere (measured by satellite) (UAH)’
- ‘Lower troposphere (measured by satellite) (RSS)’
We can also see the number of rows or records are 121 with the index starting at zero.
Since this is the first data set that we are reading and analyzing, let us check the descriptive statistics
| Year | Earth's surface | Lower troposphere (measured by satellite) (UAH) | Lower troposphere (measured by satellite) (RSS) | |
|---|---|---|---|---|
| count | 121.000000 | 121.000000 | 43.000000 | 43.000000 |
| mean | 1961.000000 | 0.267769 | 1.011395 | 1.011395 |
| std | 35.073732 | 1.015544 | 0.643309 | 0.940844 |
| min | 1901.000000 | -1.960000 | -0.202593 | -1.084286 |
| 25% | 1931.000000 | -0.410000 | 0.510657 | 0.511939 |
| 50% | 1961.000000 | 0.010000 | 1.006407 | 1.047814 |
| 75% | 1991.000000 | 0.970000 | 1.421907 | 1.664089 |
| max | 2021.000000 | 3.260000 | 2.219907 | 2.708764 |
We are using the describe() method on the DataFrame object df.
In pandas, the describe() method is used to generate descriptive statistics of the DataFrame. It provides a summary of the dataset. For example. The statistics include count, mean, standard deviation, minimum value, 25th percentile, median (50th percentile), 75th percentile, and maximum value.
By examining the output of df.describe(), we can quickly get an overview of the data in your DataFrame, including information about the range, distribution, and potential outliers.
In this case, we can immediately see
- There are 121 records or observations for Year and ‘Earth’s surface’
- We only have 43 non-missing observations (where NaN are not included) for the other two variables
- The data is from 1901 to 2021 as can be seen by the minimum and maximum
- it appears that the satellite measurements included in the variables ‘Lower troposphere (measured by satellite) (UAH)’ and ‘Lower troposphere (measured by satellite) (RSS)’ are available only recently. We can quickly check that.
| Year | Earth's surface | Lower troposphere (measured by satellite) (UAH) | Lower troposphere (measured by satellite) (RSS) | |
|---|---|---|---|---|
| count | 43.000000 | 43.000000 | 43.000000 | 43.000000 |
| mean | 2000.000000 | 1.011395 | 1.011395 | 1.011395 |
| std | 12.556539 | 1.030205 | 0.643309 | 0.940844 |
| min | 1979.000000 | -1.140000 | -0.202593 | -1.084286 |
| 25% | 1989.500000 | 0.370000 | 0.510657 | 0.511939 |
| 50% | 2000.000000 | 1.100000 | 1.006407 | 1.047814 |
| 75% | 2010.500000 | 1.625000 | 1.421907 | 1.664089 |
| max | 2021.000000 | 3.260000 | 2.219907 | 2.708764 |
In this code, we can notice that
- we first used the .dropna() that drops missing values i.e. if there is a missing value in a row (for any column), it drops the entire row
- we chained or used the .describe() method to the resulting DataFrame from df.dropna()
- we didn’t assign the resulting DataFrame to any DataFrame. The original DataFrame is not changed by these operations.
We can now see that the values for the satellite measurements included in the variables ‘Lower troposphere (measured by satellite) (UAH)’ and ‘Lower troposphere (measured by satellite) (RSS)’ are available only from 1979 to 2021 (as can be seen by the minimum and maximum value of the year variable).
In summary, so far, we have
- read the CSV data from a website link into a pandas DataFrame.
- while reading, removed the header rows and made sure that the encoding is appropriate.
- checked the data to make sure it is in line with what we expected by reading the EPA website.
Let us now create a visualization with this data. Note, we skipped the “transform data” step as the data is already in a format useful for the graph. We may need this step in the future plots.
Let’s break down the code step by step:
This line imports the plotly.express module and assigns it an alias px. This module provides a high-level interface for creating interactive plots.
This line creates a line plot using the plotly.express module. The line function is used to specify that we want to create a line plot. The df variable represents a DataFrame or a data source that contains the data we want to plot. The x parameter specifies the column in the DataFrame that will be used for the x-axis values, and the y parameter specifies the column that will be used for the y-axis values. In this case, the x-axis values are taken from the “Year” column, and the y-axis values are taken from the “Earth’s surface” column.
This line displays the plot on the screen. The show method is called on the fig object, which represents the plot that was created in the previous step.
Let us add a title and change the x-axis title to be more descriptive!
Let us start with the title.
fig = px.line(df, x= 'Year', y = "Earth's surface",
title = "Temperatures in the Contiguous 48 States, 1901–2023 "
)
fig.show()It would be nice to
- center the title of the plot
- the y-axis title is the name of the variable that we plotted. But it seems the variable should be named “Earth’s Surface Temperature” or at least the plot should include that as the y-axis title.
Let us change both! We can use update_layout method for the plotly fig object that we created.
In Plotly, the fig.update_layout() function is used to modify the layout or appearance of a plot. It allows you to customize various aspects of the plot, such as the title, axis labels, color scheme, and more.
Here is a breakdown of the fig.update_layout() function:
- fig: This is the figure object that you want to update the layout for. It represents the plot that you have created using Plotly.
- update_layout(): This is the method that allows you to update the layout of the figure.
- title_text: This parameter is used to set the title of the plot. You can provide a string value to specify the title text.
- title_x: This parameter is used to set the horizontal alignment of the title. The value should be between 0 and 1, where 0 represents left alignment and 1 represents right alignment.
- xaxis_title: change the x-axis title
- yaxis_title: change the title of the y-axis title
There are many other parameters that help you to change the appearance of the plot. We can get more details from the documentation available at https://plotly.com/python/styling-plotly-express/.
fig = px.line(df, x= 'Year', y = "Earth's surface",
title = "Temperatures in the Contiguous 48 States, 1901–2023 "
)
fig.update_layout(yaxis_title = "Earth's surface Temperature Anomaly",
title_x=0.5)
fig.show()What did we do in this code?
the update_layout method allows us to update the plot. We updated the layout of the plot by setting the y-axis title to “Earth’s surface Temperature” and adjusting the horizontal alignment of the plot title to 0.5 (which means centered).
One more final thing. When we look at the EPA graph, they have bars instead of lines. How do we do it? The nice thing with Plotly is that we need to just change the type graph.
It also would be nice to color the negative values and positive values differently as in the EPA plot.
Creating a bar graph is easy. In Plotly, all we have to change is the type of plot from px.line to px.bar.
The full code is
import pandas as pd
import plotly.express as px
url = "https://www.epa.gov/system/files/other-files/2024-06/temperature_fig-1.csv"
df = pd.read_csv(url, skiprows=6, encoding="ISO-8859-1")
fig = px.bar(df, x= 'Year', y = "Earth's surface", title = "Temperatures in the Contiguous 48 States, 1901–2023 ")
fig.update_layout(yaxis_title = "Earth's surface Temperature Anomaly",
title_x=0.5)
# Set the color of negative values to red and positive values to blue
fig.update_traces(marker_color=['blue' if val < 0 else 'red' for val in df["Earth's surface"]])
fig.show()We first updated the line plot to bar plot.
Then, we used the update_traces() method to update the properties of the traces in the plot. A trace is a set of data points that are plotted together. In this case, it is updating the marker color of the traces.
The marker_color parameter is used to specify the color of the markers in the plot. It expects a list of colors, where each color corresponds to a data point in the plot.
The color of each marker is determined by the list comprehension [‘blue’ if val < 0 else ‘red’ for val in df[“Earth’s surface”]]. This list comprehension iterates over the values in the “Earth’s surface” column of the df DataFrame and assigns the color ‘blue’ if the value is less than 0, and ‘red’ otherwise. We chose red for hotter temperatures!
As you can see, we get a graph similar to what’s on the EPA’s website. But here we have only used one variable “Earth’s surface” whereas the EPA also include data from the satellite measurements where available (from 1979 onwards).
For now, we are not going to add them to keep the plot simple in this introductory chapter. We will include code for plotting on two axes and more than one variable later in the book.
Isn’t it fun coding it and making our own though?
But wait! Why did we code it? Our goal was not to just code and show a plot. But through the graph understand what is happening with the climate.
First, what exactly does the indicator measure? From the EPA website.
This indicator examines U.S. and global surface temperature patterns over time. U.S. surface measurements come from weather stations on land, while global surface measurements also incorporate observations from buoys and ships on the ocean, thereby providing data from sites spanning much of the surface of the Earth. This indicator starts at 1901 except for the detailed map of Alaska, where reliable statewide records are available back to 1925. For comparison, this indicator also displays satellite measurements that can be used to estimate the temperature of the Earth’s lower atmosphere since 1979.
This indicator shows annual anomalies, or differences, compared with the average temperature from 1901 to 2000. For example, an anomaly of +2.0 degrees means the average temperature was 2 degrees higher than the long-term average. Anomalies have been calculated for each weather station. Daily temperature measurements at each site were used to calculate monthly anomalies, which were then averaged to find an annual temperature anomaly for each year. Anomalies for the contiguous 48 states and Alaska have been determined by calculating average anomalies for areas within each state based on station density, interpolation, and topography. These regional anomalies are then averaged together in proportion to their area to develop national results. Similarly, global anomalies have been determined by dividing the world into a grid, averaging the data for each cell of the grid, and then averaging the grid cells together.
As the plot clearly shows, average temperatures are increasing over time. As the EPA website states, the plot shows that:
- Since 1901, the average surface temperature across the contiguous 48 states has risen at an average rate of 0.17°F per decade.
- Average temperatures have risen more quickly since the late 1970s (0.32 to 0.51°F per decade since 1979).
- For the contiguous United States, nine of the 10 warmest years on record have occurred since 1998, and 2012 and 2016 were the two warmest years on record.
But before that, let us recreate the graphs slightly differently.
- let us introduce functions which take some parameters
- introduce headers to mimic the behavior of an internet browser. Sometimes, EPA websites or others give an error because they want to make sure that a valid request is coming through. The headers would help with it.
- also add the two other variables in the dataset and the EPA plot that we didn’t plot so far.
Functions: Functions are useful for making the code reproducible and to keep the code modular. If we have to repeat the same piece of code, only changing the URL, we can either:
- cut and copy the entire code and change the URL or
- write a function that takes the URL as the input and call with the new URL
Let us read the data for the figure using a function.
from io import StringIO
import pandas as pd
import requests
def url_data_request(url, skiprows):
headers = {
"User-Agent": "Mozilla/5.0 (X11; Ubuntu; Linux x86_64; rv:109.0) Gecko/20100101 Firefox/118.0",
}
f = StringIO(requests.get(url, headers=headers).text)
df_url = pd.read_csv(f, skiprows = skiprows)
return df_urlLet’s break down the code step-by-step:
We use a Python function called url_data_request that takes two parameters: url and skiprows. This function is designed to retrieve data from a specified URL and return it as a pandas DataFrame.
The function begins by importing the necessary modules: StringIO from the io module, pandas as pd, and requests.
The url_data_request function definition starts with the function name and the two parameters it expects: url and skiprows.
Inside the function, a dictionary called headers is created. This dictionary contains a user-agent string that is used to identify the client making the request. In this case, it is set to mimic the behavior of the Mozilla Firefox browser.
The next line of code creates a StringIO object called f. StringIO is a class that allows you to treat a string as a file-like object. In this case, it is used to store the text content of the response from the specified URL.
The requests.get() function is called with the url and headers as arguments. This function sends an HTTP GET request to the specified URL and returns a response object.
The text attribute of the response object is accessed using dot notation (requests.get(…).text) to retrieve the text content of the response.
The text content is then passed as an argument to the StringIO object f, effectively storing the response text as if it were a file.
The next line of code creates a pandas DataFrame called df_url by reading the CSV data from the f object. The pd.read_csv() function is used for this purpose. The skiprows parameter is passed to the function to specify the number of rows to skip at the beginning of the CSV file.
Finally, the function returns the df_url DataFrame.
This function can be used to easily retrieve data from a specified URL and convert it into a pandas DataFrame for further analysis or manipulation. It is important to note that the function assumes the data at the specified URL is in CSV format. Additionally, the skiprows parameter allows you to skip a certain number of rows if needed, which can be useful if the CSV file contains header information or other irrelevant data at the beginning.
Why did we need this? In case, a different file has 8 rows of headers, all we need to do is change the skiprows parameter. The same thing with URL. Functions can be quite handy and we will make extensive use of them later in the book. They help us write short and clear code as we can reuse code and make it more readable.
Let us start with the U.S. first. Let us use the function that we created and get the data.
url = "https://www.epa.gov/system/files/other-files/2024-06/temperature_fig-1.csv"
df = url_data_request(url, 6)
df| Year | Earth's surface | Lower troposphere (measured by satellite) (UAH) | Lower troposphere (measured by satellite) (RSS) | |
|---|---|---|---|---|
| 0 | 1901 | -0.15 | NaN | NaN |
| 1 | 1902 | -0.43 | NaN | NaN |
| 2 | 1903 | -1.40 | NaN | NaN |
| 3 | 1904 | -0.86 | NaN | NaN |
| 4 | 1905 | -1.02 | NaN | NaN |
| ... | ... | ... | ... | ... |
| 118 | 2019 | 0.66 | 1.133593 | 1.221429 |
| 119 | 2020 | 2.35 | 1.970593 | 2.290629 |
| 120 | 2021 | 2.49 | 2.059093 | 2.485329 |
| 121 | 2022 | 1.37 | 1.567093 | 1.596879 |
| 122 | 2023 | 2.39 | NaN | 1.886229 |
123 rows × 4 columns
It is always a good idea to first check the data structure again by listing the columns and information on the number of observations.
| Year | Earth's surface | Lower troposphere (measured by satellite) (UAH) | Lower troposphere (measured by satellite) (RSS) | |
|---|---|---|---|---|
| count | 123.000000 | 123.000000 | 44.000000 | 45.000000 |
| mean | 1962.000000 | 0.294472 | 1.050889 | 1.050889 |
| std | 35.651087 | 1.029710 | 0.641933 | 0.919074 |
| min | 1901.000000 | -1.960000 | -0.174407 | -1.053021 |
| 25% | 1931.500000 | -0.410000 | 0.542593 | 0.557979 |
| 50% | 1962.000000 | 0.010000 | 1.037593 | 1.078179 |
| 75% | 1992.500000 | 1.090000 | 1.480468 | 1.749279 |
| max | 2023.000000 | 3.260000 | 2.252593 | 2.677329 |
The data seems to be from 1901-2023 with some of the variables measured only for the last four decades or so.
import plotly.express as px
fig = px.bar(df, x = 'Year',
y = ["Earth's surface",
'Lower troposphere (measured by satellite) (UAH)',
'Lower troposphere (measured by satellite) (RSS)'])
fig.update_layout(
yaxis_title="Temperature Anomaly (degree FH)",
title='Temperature Anomalies U.S., 1901–2021 ',
hovermode="x",
legend=dict(x=0.4,y=1),
title_x = 0.5
)
fig.update_layout(legend={'title_text':''})
fig.show()We used “Earth’s surface” for “Earth’s Surface” as Earth’s already has a
We can see that the temperature over the past few years has been much hotter than the previous hundred years. To see it more clearly, let us list the years with the highest temperatures.
| Year | Earth's surface | |
|---|---|---|
| 111 | 2012 | 3.26 |
| 115 | 2016 | 2.90 |
| 116 | 2017 | 2.53 |
| 120 | 2021 | 2.49 |
| 122 | 2023 | 2.39 |
| 114 | 2015 | 2.38 |
| 119 | 2020 | 2.35 |
| 105 | 2006 | 2.23 |
| 97 | 1998 | 2.21 |
| 33 | 1934 | 2.08 |
As we can see, in the U.S. 2012 is the hottest year. Most importantly, 9 of the top 10 hottest years since 1901 were 1998 and afterwards, with most of them 2015 or after. Only 1934 makes it to the top 10.
There are two other figures at the EPA website for temperature. We can quickly plot both using the same code with minor modifications.
5.1.1 Temperatures Worldwide
We can recreate 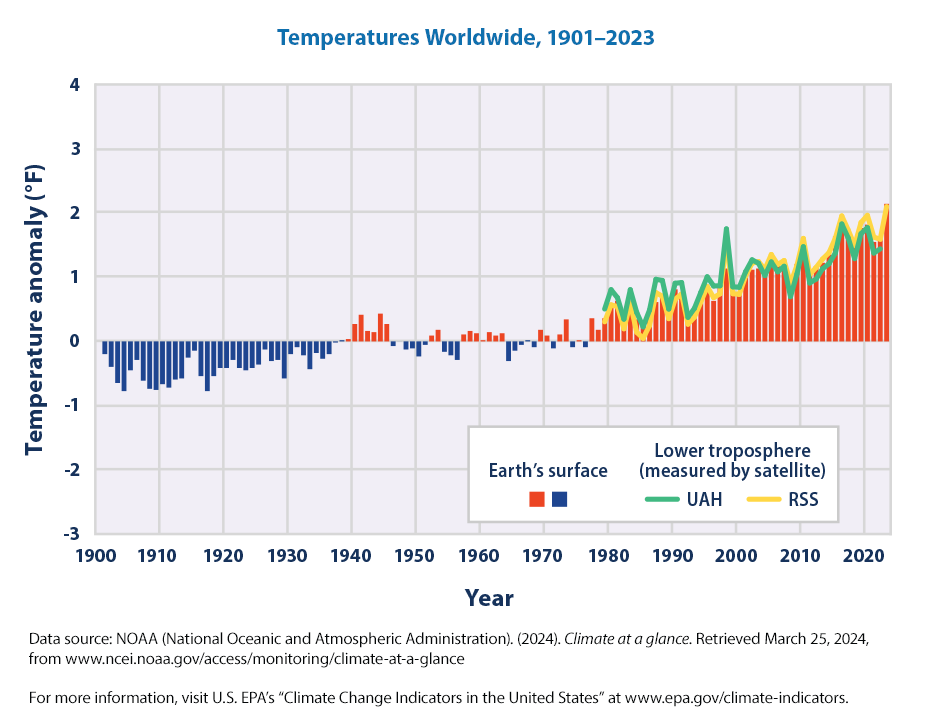
We can use our function and just change the URL
url = "https://www.epa.gov/system/files/other-files/2024-06/temperature_fig-2.csv"
df = url_data_request(url, 6)
df| Year | Earth's surface (land and ocean) | Lower troposphere (measured by satellite) (UAH) | Lower troposphere (measured by satellite) (RSS) | |
|---|---|---|---|---|
| 0 | 1901 | -0.198 | NaN | NaN |
| 1 | 1902 | -0.396 | NaN | NaN |
| 2 | 1903 | -0.648 | NaN | NaN |
| 3 | 1904 | -0.774 | NaN | NaN |
| 4 | 1905 | -0.450 | NaN | NaN |
| ... | ... | ... | ... | ... |
| 118 | 2019 | 1.764 | 1.675955 | 1.849987 |
| 119 | 2020 | 1.818 | 1.770455 | 1.962187 |
| 120 | 2021 | 1.548 | 1.375955 | 1.623037 |
| 121 | 2022 | 1.620 | 1.440455 | 1.590787 |
| 122 | 2023 | 2.142 | NaN | 2.098687 |
123 rows × 4 columns
Here, we read the dataset again and skip the first 6 rows. We will probably be doing this in all the datasets this chapter as EPA lists information about the data in the first six rows.
You will notice that the data structure is the same, but now the variable name is slightly different, “Earth’s surface (land and ocean)” instead of just “Earth’s surface” before.
How did we know? Honestly, we tried to run the same code, got an error, and realized that the variable name has changed and we need to make minor changes to the code.
That’s why it is always good to have a quick look at the data before running the code.
Notice below that the only line we changed was the URL! That’s the beauty of programming. Once we have the code, for most of the time, if there are no breaking changes (URL change, data format change, etc.), we can
- create new plots with minor changes in the code. In this case,
- URL,
- title, and
- the variable name
- continuously update the plots
import pandas as pd
import plotly.express as px
url = "https://www.epa.gov/system/files/other-files/2024-06/temperature_fig-2.csv"
df = url_data_request(url, 6)
fig = px.bar(df, x= 'Year', y = "Earth's surface (land and ocean)", title = "Temperatures Worldwide, 1901–2023")
fig.update_layout(yaxis_title = "Earth's surface (land and ocean)",
title_x=0.5)
# Set the color of negative values to red and positive values to blue
fig.update_traces(marker_color=['blue' if val < 0 else 'red' for val in df["Earth's surface (land and ocean)"]])
fig.show()What are the insights? Again, taking verbatim from the EPA website
Worldwide, 2023 was the warmest year on record, 2016 was the second-warmest year, and 2014–2023 was the warmest decade on record since thermometer-based observations began. Global average surface temperature has risen at an average rate of 0.17°F per decade since 1901 (Figure 2), similar to the rate of warming within the contiguous United States. Since the late 1970s, however, the United States has warmed faster than the global rate.
From both these graphs, U.S. and worldwide, the main takeaways are:
- Since 1901, the average surface temperature across the globe and across the contiguous 48 states has risen at an average rate of 0.17°F per decade
- Since the late 1970s, the United States has warmed faster than the global rate.
5.1.2 Temperatures Across the USA
Let us recreate the third figure from the EPA Climate Indicators on Temperature.
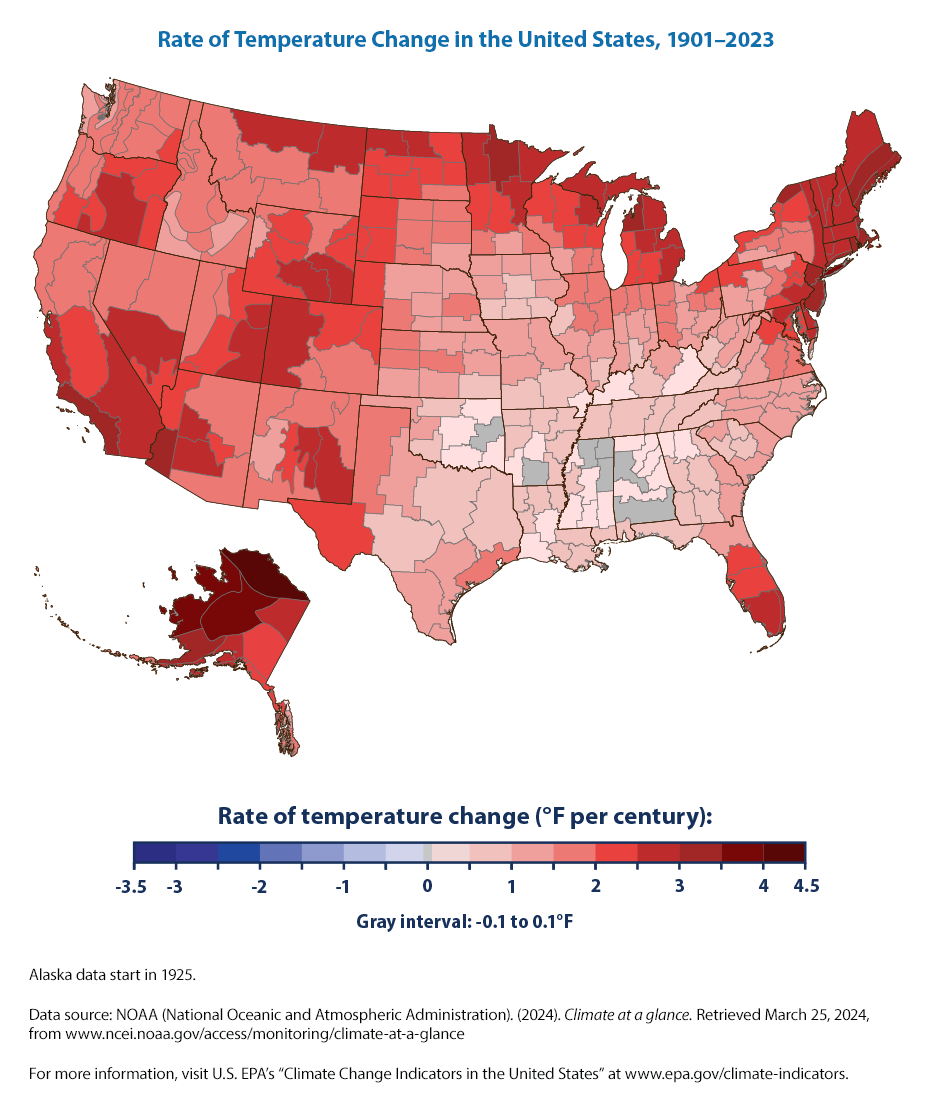
Now, this is a different plot from the line and bar plots that we have seen so far. It is showing temperature change over the past century across the U.S. We need to find a way to plot it on the U.S. map.
We need to expand our repertoire to include choropleth maps!
What is a choropleth map?
A choropleth map is a type of thematic map that uses color shading or patterns to represent data values for specific geographic areas, such as countries, states, or regions. It is commonly used to visualize data that is associated with different locations.
The term “choropleth” comes from the Greek words “choros” meaning area or region, and “plethos” meaning value or quantity. In a choropleth map, each geographic area is filled with a color or pattern that corresponds to a particular data value or range of values.
To create a choropleth map, you typically start with a map of the geographic areas you want to represent. Each area is then assigned a color or pattern based on the data value it represents. The color or pattern intensity can be used to indicate the magnitude or density of the data.
For example, let’s say you have a dataset that contains the average temperature of different cities in a country. You can create a choropleth map where each city is represented by a polygon on the map, and the color of the polygon corresponds to the average temperature of that city. Warmer cities can be represented with a darker color, while cooler cities can be represented with a lighter color.
Choropleth maps are useful for visualizing spatial patterns and variations in data across different geographic areas. They can help identify trends, patterns, and disparities in the data. However, it’s important to choose appropriate color schemes and classification methods to ensure the map accurately represents the data and is easy to interpret.
We are going to use Plotly to generate the choropleth map to recreate a plot similar to the EPA’s temperature change across the U.S.
Let us get the data for this figure first and check the variable names.
#figure 3 rate of temperature change in US
import pandas as pd
url = "https://www.epa.gov/system/files/other-files/2022-07/temperature_fig-3.csv"
df = pd.read_csv(url,skiprows = 6, encoding = "ISO-8859-1")
df| Climate Division | Temperature Change | |
|---|---|---|
| 0 | 101 | 0.169506 |
| 1 | 102 | 0.229452 |
| 2 | 103 | -0.206860 |
| 3 | 104 | 0.277881 |
| 4 | 105 | 0.220086 |
| ... | ... | ... |
| 339 | 4806 | 2.149714 |
| 340 | 4807 | 2.063914 |
| 341 | 4808 | 2.676568 |
| 342 | 4809 | 2.091457 |
| 343 | 4810 | 2.726776 |
344 rows × 2 columns
We changed the URL and read the data into a dataframe. As before, we have skipped the first six rows where EPA was puttimg some header information. We also used the appropriate encoding to read the data.
Note that the DataFrame df now holds the data or figure 3, the data for temperature changes across the U.S. The previous data in the dataframe is over written or lost.
It seems that the DataFrame has only two variables
- ‘Climate Division’,
- ‘Temperature Change’
There are 344 rows and 2 columns.
How do we map the climate divisions into the U.S. map?
We need a cross walk that can map between climate divisions and the U.S. map.
First, let us get the climate divisions data from NOAA.
by the way, if you see an error below, it is because the National Center for Environmental Information (NCEI) that hosts the data was hit by Hurricane Helene and the data center is down. It is ironical but highlights the reality of climate change and extreme weather. 
import pandas as pd
url = "https://www.ncei.noaa.gov/pub/data/cirs/climdiv/county-to-climdivs.txt"
df_div= pd.read_csv(url, skiprows = 3,sep = ' ')
df_div| POSTAL_FIPS_ID | NCDC_FIPS_ID | CLIMDIV_ID | |
|---|---|---|---|
| 0 | 1033.0 | 1033.0 | 101.0 |
| 1 | 1059.0 | 1059.0 | 101.0 |
| 2 | 1077.0 | 1077.0 | 101.0 |
| 3 | 1079.0 | 1079.0 | 101.0 |
| 4 | 1083.0 | 1083.0 | 101.0 |
| ... | ... | ... | ... |
| 3103 | 56031.0 | 48031.0 | 4808.0 |
| 3104 | 56013.0 | 48013.0 | 4809.0 |
| 3105 | 56001.0 | 48001.0 | 4810.0 |
| 3106 | 56007.0 | 48007.0 | 4810.0 |
| 3107 | NaN | NaN | NaN |
3108 rows × 3 columns
Let’s break down the code step by step:
The url variable is assigned the value “https://www.ncei.noaa.gov/pub/data/cirs/climdiv/county-to-climdivs.txt”. This URL points to a CSV file that contains climate division crosswalk to other geograhic level variables such as FIPS.
The pd.read_csv() function is called to read the CSV file. This function is provided by the pandas library and is commonly used to read tabular data from CSV files into a DataFrame. It takes several parameters:
url: The URL of the CSV file to be read.
skiprows: The number of rows to skip from the beginning of the file. In this case, skiprows = 3 indicates that the first 3 rows of the file will be skipped.
sep: The delimiter used to separate values in the CSV file. In this case, sep = ’ ’ indicates that a space character is used as the delimiter.
The resulting DataFrame is assigned to the variable df_div. A DataFrame is a two-dimensional tabular data structure provided by the pandas library. It organizes data in rows and columns, similar to a table in a spreadsheet.
Finally, the df_div DataFrame is displayed.
Overall, this code retrieves a CSV file from a specific URL, skips the first 3 rows, and stores the remaining data in a DataFrame for further analysis or manipulation.
Why do we need this crosswalk and the FIPS code? First, let us check what FIPS code is. You can get the full list from FCC website
The FIPS code, also known as the Federal Information Processing Standards code, is a unique identifier assigned to geographic areas in the United States. It is used by various government agencies, including the U.S. Census Bureau and the National Weather Service, to identify and organize data related to specific locations.
The FIPS code consists of a five-digit number, where the first two digits represent the state or territory, and the last three digits represent the county or county equivalent within that state. For example, in the FIPS code “06037”, the first two digits “06” represent California, and the last three digits “037” represent Los Angeles County.
Before combining two datasets, we need to check on a couple of issues first
- is there a common variable in both datasets?
- are the variables that we are merging on of the same data type?
We can check by running
We can see that:
- The Climate Division Id is listed as “Climate Division” in df dataset and as “CLIMDIV_ID” in the crosswalk df_div dataset. This is a common occurence as data comes from different organizations. Even within the same organization, the variable names may change over time. We don’t need to necesssarily change the name of the variable to merge these two datasets.
- “Climate Division” is int64 data type and “CLIMDIV_ID” is float64 datatype (it has a decimal)
For transparency, let us change the name of the variable in one of the datasets so that there is a common variable in both the datasets.
We could have done the same in three steps instead of chaining the methods.
This line of code is using the rename() function to rename a column in the DataFrame. The columns parameter is a dictionary that specifies the old column name as the key and the new column name as the value.
What it means is that we can rename multiple variables if needed by listing them in the dictionary. In this case, the code is renaming only one column with the old name “CLIMDIV_ID” to the new name “Climate Division”.
.dropna(): This method is called on the DataFrame after renaming the column. It removes any rows that contain missing or NaN (Not a Number) values. The dropna() function is commonly used to handle missing data in a DataFrame.
.astype(int): This method is called on the DataFrame after dropping the rows with missing values. It converts the data type of the DataFrame to integer (int). The astype() function is used to change the data type of a column or the entire DataFrame.
Instead of writing three lines of code and assigning the result of each operation to the same dataset, it is easier to chain all three methods in one line of code
In Python, method chaining refers to the practice of calling multiple methods or functions on the same object in a single line of code. This allows you to perform multiple operations on an object without the need for intermediate variables.
The dot (.) operator is used to chain methods or functions together. Each method or function is called on the result of the previous method or function, and the output of each call becomes the input for the next call.
In this case, the output of df_div.rename(columns ={“CLIMDIV_ID”: “Climate Division”}) is the input for .dropna() and it in turn is the input of .astype(int).
Before we combine two datasets, it is also a good idea to check the number of observations in each dataset so that we can be confident that the data are combined correctly.
The number of observations in the datasets can be listed with the following command.
The shape attribute of a DataFrame returns a tuple containing the number of rows and columns in the DataFrame.
So, in this code, df.shape is used to retrieve the shape of the DataFrame df, and the values of the number of rows and columns are assigned to the variables r and c, respectively.
We can see that df has 344 number of observations.
Similarly,
We can see that df_div has 3107 number of observations. Also note that we over wrote the values of r, c as we don’t need them later.
Let us make sure that there are no duplicates in each dataset.
It appears that there are no duplicate observations (where all the values of all the variables are the same). We still have multiple observations for each Climate Division as there can be many FIPS within each Climate Division.
There are 344 climate divisions across teh U.S. where as the crosswalk file df_div shows that there are more than 3000 FIPS in the U.S.
Now we can merge with our EPA dataset based on a common variable Climate Division.
| POSTAL_FIPS_ID | NCDC_FIPS_ID | Climate Division | Temperature Change | |
|---|---|---|---|---|
| 0 | 1033 | 1033 | 101 | 0.169506 |
| 1 | 1059 | 1059 | 101 | 0.169506 |
| 2 | 1077 | 1077 | 101 | 0.169506 |
| 3 | 1079 | 1079 | 101 | 0.169506 |
| 4 | 1083 | 1083 | 101 | 0.169506 |
| ... | ... | ... | ... | ... |
| 3102 | 56025 | 48025 | 4808 | 2.676568 |
| 3103 | 56031 | 48031 | 4808 | 2.676568 |
| 3104 | 56013 | 48013 | 4809 | 2.091457 |
| 3105 | 56001 | 48001 | 4810 | 2.726776 |
| 3106 | 56007 | 48007 | 4810 | 2.726776 |
3107 rows × 4 columns
Let us understand the merge better as this is an important method and we will use it many times in the book to combine various datasets. Let’s break down the code step-by-step:
The first line of code initializes a new DataFrame called df_merge by merging two existing dataframes, df_div and df, using the merge() function.
The merge() function is used to combine two dataframes based on a common column or index. In this case, the common column is specified as ‘Climate Division’ using the on parameter.
The how parameter is set to ‘inner’, which means that only the rows with matching values in the ‘Climate Division’ column will be included in the merged dataframe. Other possible values for the how parameter include ‘left’, ‘right’, and ‘outer’, which determine how the merge handles non-matching rows.
Finally, the merged dataframe df_merge is printed to the output.
The combined or merged dataset has
- 3107 observations as in df_div
- an additional column, “Temperature Change” from df is added to the columns in df_div
Why did we want to get the FIPS code? So, we can use the FIPS code to create a chloropeth map of the U.S.
But wait! There is one more data wrangling task that needs to be done first!
FIPS should be 5 digits but the leading zero is missing as it is in a float64 format. We we will also see the geojson file that we need to combine with to plot a map has FIPS as a string variable. So, we need to change it.
df_merge['POSTAL_FIPS_ID'] = df_merge.POSTAL_FIPS_ID.astype(str).str.pad(5, fillchar='0')
df_merge.head()| POSTAL_FIPS_ID | NCDC_FIPS_ID | Climate Division | Temperature Change | |
|---|---|---|---|---|
| 0 | 01033 | 1033 | 101 | 0.169506 |
| 1 | 01059 | 1059 | 101 | 0.169506 |
| 2 | 01077 | 1077 | 101 | 0.169506 |
| 3 | 01079 | 1079 | 101 | 0.169506 |
| 4 | 01083 | 1083 | 101 | 0.169506 |
Let’s break down the chained code step-by-step:
df_merge[‘POSTAL_FIPS_ID’]: This part of the code is accessing a column named ‘POSTAL_FIPS_ID’ in the DataFrame df_merge.
.astype(str): This method is used to convert the values in the ‘POSTAL_FIPS_ID’ column to strings. It ensures that all the values in the column are treated as strings.
.str.pad(5, fillchar=‘0’): This method is applied to each string value in the ‘POSTAL_FIPS_ID’ column. It pads each string with leading zeros (‘0’) to make it a total of 5 characters long. This is achieved using the pad method of the string accessor (str).
For example, if the original value in the ‘POSTAL_FIPS_ID’ column is ‘123’, after applying this method, it will become ‘00123’.
- Finally, the result of the above operations is assigned back to the ‘POSTAL_FIPS_ID’ column in the df_merge DataFrame.
In summary, this code snippet converts the values in the ‘POSTAL_FIPS_ID’ column of the df_merge DataFrame to strings and pads them with leading zeros to make them 5 characters long.
GeoJSON format: GeoJSON is a format for encoding a variety of geographic data structures. More information can be found at https://geojson.org/ and https://plotly.com/python/choropleth-maps/
Before mapping over the U.S. map, it is a good idea to understand how temperature has changed since 1901.
| NCDC_FIPS_ID | Climate Division | Temperature Change | |
|---|---|---|---|
| count | 3107.000000 | 3107.000000 | 3107.000000 |
| mean | 24888.250724 | 2482.915996 | 1.345643 |
| std | 13713.450404 | 1369.327474 | 0.782315 |
| min | 1001.000000 | 101.000000 | -0.206860 |
| 25% | 13046.000000 | 1302.500000 | 0.711382 |
| 50% | 23213.000000 | 2305.000000 | 1.218715 |
| 75% | 39008.000000 | 3901.000000 | 1.874695 |
| max | 48045.000000 | 4810.000000 | 3.585281 |
We can see that the average change in temperature for the contiguous united states is 1.34 degree Fahrenheit since 1901. The maximum is 3.58 degrees. There are very few climate divisions with a negative value for temperature change i.e. there are few with a net decrease in temperature.
Lets plot a histogram of the temperature changes so that we can see more visually these changes. We need a bit of code for this plot! Let us take it step-by-step
As before, we have
- plotly.express is imported as px for creating interactive plots.
- pandas is imported as pd for data manipulation and analysis.
Next, we wanted to create a new column
- A new column Change is added to the DataFrame df
- The apply function is used to classify each value in the Temperature Change column.
- If the temperature change is less than 0, it is classified as ‘Decrease’; otherwise, it is classified as ‘Increase’.
The call out note below gives more details about apply and lamda expressions.
Lambda functions: Lamda functions also known as anonymous functions, are small, unnamed functions defined using the lambda keyword. They are often used for short, simple operations that are not reused elsewhere in the code. In this case, we used the lamda function to create Decrease / Increase value for the Change variable based on whether the Temperature Change is Negative or Positive.
apply: The apply method is used to apply a function along an axis of the DataFrame or Series.
Here’s a detailed explanation:
apply Method in Pandas The apply method can be used on both DataFrame and Series objects. It allows you to apply a function to each element, row, or column of the DataFrame or Series.
fig = px.histogram(df_merge, x="Temperature Change", color="Change",
color_discrete_map={'Decrease': 'blue', 'Increase': 'red'},
nbins=20)- A histogram is created using plotly.express.histogram.
- The x-axis represents the Temperature Change.
- The bars are colored based on the Change column, with ‘Decrease’ in blue and ‘Increase’ in red.
- The nbins parameter is set to 20, specifying the number of bins in the histogram.
Next, we want to update the layout similar to the line plot that we did before.
fig.update_layout(
title="Rate of Temperature Change in the United States, 1901–2023",
xaxis_title="Temperature Change",
yaxis_title="Number of FIPS",
title_x = 0.5,
legend=dict(
x=0.8,y=0.95
)
)- The layout of the plot is updated to include a title and axis labels.
- title_x=0.5 centers the title.
- The legend for “Change” is positioned at x=0.8 (80% from the left) and y=0.95 (95% from the bottom).
The full code to create the plot is
import plotly.express as px
import pandas as pd
# Add a column to classify the data as 'Negative' or 'Positive'
df_merge['Change'] = df_merge['Temperature Change'].apply(lambda x: 'Decrease' if x < 0 else 'Increase')
# Create the histogram
fig = px.histogram(df_merge, x="Temperature Change", color="Change",
color_discrete_map={'Decrease': 'blue', 'Increase': 'red'},
nbins=20)
# Update the layout
fig.update_layout(
title="Rate of Temperature Change in the United States, 1901–2023",
xaxis_title="Temperature Change",
yaxis_title="Number of FIPS",
title_x = 0.5,
legend=dict(
x=0.8, # X position (0 is far left, 1 is far right)
y=0.95)
)
# Show the plot
fig.show()As the EPA keypoint for this measure states:
Since 1901, the average surface temperature across the contiguous 48 states has risen at an average rate of 0.17°F per decade … Average temperatures have risen more quickly since the late 1970s (0.32 to 0.51°F per decade since 1979). For the contiguous United States, nine of the 10 warmest years on record have occurred since 1998, and 2012 and 2016 were the two warmest years on record.
The plot clearly shows that almost all of the U.S. experienced an incerase in temperature and very few experienced a decrease in temperature.
5.1.2.1 Choropleth Map
So far, we have presented the data without using the geographic information. Remember, we started this exercise to plot the temperature changes on the U.S. map. We haven’t done that yet!
We followed the code and tutorial from Plotly Choropleth
What is the code doing?
- As before, we imported plotly.express
- Next, the code imports the urlopen function from the urllib.request module. This function allows us to open URLs and retrieve data from them.
- After that, the code imports the json module. This module provides functions for working with JSON data.
- The code then uses the with statement to open a URL using the urlopen function. The URL in this case is ‘https://raw.githubusercontent.com/plotly/datasets/master/geojson-counties-fips.json’. This URL points to a JSON file containing the geographic data for counties in the United States.
Now that we have the coordinates for each of the FIPS code, we can plot temperature changes over the U.S. map.
fig = px.choropleth(df_merge, geojson=counties, locations='POSTAL_FIPS_ID', color='Temperature Change',
color_continuous_scale=[(0, 'green'), (0.025, 'grey'), (0.50, 'yellow'), (1, 'red')],
range_color=(-0.25, 3.75),
scope="usa",
labels={'Temperature Change': 'Δ°F'}
)
fig.update_layout(margin={"r":0,"t":0,"l":0,"b":0})
fig.show()Let us go step-by-step up as this is the first time that we are plotting this type of map.
The components that we have used are
px.choropleth: This function creates a choropleth map, which is a type of map where areas are shaded in proportion to the value of a variable.
df_merge: This is the DataFrame containing the data to be plotted.
geojson=counties: This specifies the GeoJSON data that defines the geographical boundaries of the counties.
locations=‘POSTAL_FIPS_ID’: This specifies the column in df_merge that contains the FIPS codes, which are used to match the data to the geographical areas.
color=‘Temperature Change’: This specifies the column in df_merge that contains the values to be used for coloring the map.
color_continuous_scale=[(0, ‘green’), (0.025, ‘grey’), (0.50, ‘yellow’), (1, ‘red’)]: This defines the color scale for the map. The scale is continuous and maps values to colors. For example, values close to 0 are green, values around 0.025 are grey, values around 0.50 are yellow, and values close to 1 are red. range_color=(-0.25, 3.75): This sets the range of values for the color scale. Values below -0.25 will be clipped to -0.25, and values above 3.75 will be clipped to 3.75. How did we come up with these numbers - frankly, by plotting without using these colors and finding that the resulting plot doesn’t look good - the issue is that there are very few negative values i.e. decreases in temperature. Only around 2.5% of the sample. So, the default color scheme doesn’t look good. So, we chose to put the lowest values (negative and close to zero) as green and then transition to gery and then yellow and finally for the large increases in temperature, red - the range is the range of temperature increases that we saw through the .describe() method before
scope=“usa”: This limits the map to the USA.
labels={‘Temperature Change’: ‘Temperature Change’}: This sets the label for the color bar. Updating Layout:
fig.update_layout: This function updates the layout of the figure. margin={“r”:0,“t”:0,“l”:0,“b”:0}: This sets the margins of the plot to zero on all sides (right, top, left, bottom), effectively removing any padding around the plot.
fig.show: This function displays the plot in an interactive window.
In summary, his code creates and displays a choropleth map of the USA, where each county is colored based on the ‘Temperature Change’ value from the df_merge DataFrame. The color scale ranges from green to red, with intermediate colors grey and yellow, and the map is displayed with no margins around it.
Let us do the choropleth map slightly differently. The EPA uses a gray interval from -0.1 °F to +0.1 °F.
Let us create a new variable that takes three values: negative (< -0.1°F), near zero (-0.1 °F to +0.1 °F) and positive (> 0.1°F)
We can do that by using pd.cut.
What did we do using pd.cut? This function is used to categorize the continuous Change values into discrete bins:
- bins=[-5, -0.01, 0.01, 5] will create three bins (-5, -0.01), (-0.01, 0.01) and (0.01, 5). We chose these bins based on the distribution of Temperature Change
- the corresponding labels created will be
- (-5, -0.01): Negative
- (-0.01, 0.01) : Near Zero
- (0.01, 5) : Positive
We can modify our previous choropleth map code slightly
import plotly.express as px
import pandas as pd
fig = px.choropleth(
df_merge,
geojson=counties,
locations='POSTAL_FIPS_ID',
color='Category',
color_discrete_map={'Negative': 'blue', 'Near Zero': 'gray', 'Positive': 'red'},
scope="usa",
labels={'Category': 'Temperature Change'}
)
fig.update_layout(
margin={"r":0,"t":0,"l":0,"b":0}
)
fig.show()We modified:
- color_discrete_map: Maps each category to a specific color: blue for ‘Negative’, grey for ‘Near Zero’, and red for ‘Positive’.
- color=‘Category’: Tells Plotly to use the categorized Category column for coloring the map.
This will generate a choropleth map where negative changes are colored blue, near-zero changes are grey, and positive changes are red.
Now the map looks similar to the EPA map. Note that our FIPS code has only contiguous U.S. states so, we are missing Hawaii.
We can immediately see that the entire contiguous U.S. state map is red i.e. the temperature change is more than > 0.1°F! We are not suprised as the mean and median temperature based on the descriptive stats are both positive and there are very few negative values. We can see these few FIPS with a decline in temperature.
As the EPA states about this figure > Some parts of the United States have experienced more warming than others. The North, the West, and Alaska have seen temperatures increase the most, while some parts of the Southeast have experienced little change. Not all of these regional trends are statistically significant, however.
5.2 Summary
We have covered a lot of ground so far in this chapter. Let us summarize.
5.2.1 Programming
5.2.1.1 Data Manipulation with Pandas:
- DataFrame Creation: Creating DataFrames from various CSV files downloaded from internet.
- Filtering and Selecting Data: Using conditions to filter rows and selecting specific columns.
- Aggregation and Grouping: Summarizing data using groupby operations.
- Merging and Joining: Combining multiple DataFrames using merge or join operations.
- Handling Missing Data: Techniques for dealing with NaN values, such as filling or dropping them. Data Visualization with Plotly:
5.2.1.2 Creating Plots Using Plotly
- Generating different types of plots (e.g., line plots, scatter plots, bar charts).
- Customizing Plots: Modifying plot attributes like titles, axis labels, legends, and colors.
- Creating a Choropleth map.
5.2.1.3 String Operations:
- Manipulating string data to clean and standardize text fields.
- Date and Time Handling: Parsing and formatting date and time data for analysis.
- Data Transformation: Applying transformations to data, such as scaling or encoding categorical variables.
5.3 Climate Change Insights:
We can reprint the key points that EPA highlights for these figures.
Since 1901, the average surface temperature across the contiguous 48 states has risen at an average rate of 0.17°F per decade (Figure 1). Average temperatures have risen more quickly since the late 1970s (0.32 to 0.51°F per decade since 1979). For the contiguous United States, nine of the 10 warmest years on record have occurred since 1998, and 2012 and 2016 were the two warmest years on record.
Worldwide, 2023 was the warmest year on record, 2016 was the second-warmest year, and 2014–2023 was the warmest decade on record since thermometer-based observations began. Global average surface temperature has risen at an average rate of 0.17°F per decade since 1901 (Figure 2), similar to the rate of warming within the contiguous United States. Since the late 1970s, however, the United States has warmed faster than the global rate.
Some parts of the United States have experienced more warming than others (Figure 3). The North, the West, and Alaska have seen temperatures increase the most, while some parts of the Southeast have experienced little change. Not all of these regional trends are statistically significant, however.